CRISPR meets the Digenome
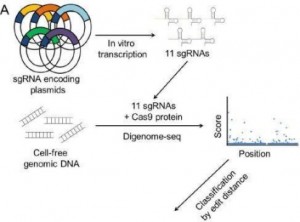
Multiplex Digenome Seq
Thinking about the prospects of CASPR-Cas9 gene editing might bring to mind the title of Irving Stone’s biography of Michelangelo: “The Agony and the Ecstasy.” Gene editing has enormous promise, but it is now limited by cutting out genes at off-target sites. This is a problem of the targeting RNAs that guide the endonuclease to its intended site. The fewer number of RNA bases, the greater the likelihood the Cas9 will act on dozens, hundreds or even thousands of sites. New, longer targeting sequences have been discovered in various bacteria, but off targeting remains a limiting factor. Researchers at the Institute for Basic Science (Daejeon, South Korea), however, have developed a DNA sequencing technique called “Digenome-seq” that searches for CRISPR’s targeting sites in cell free genomic DNA. “Dig” indicates digested DNA, and their technique resembles Craig Venter’s “shotgun sequencing” used in the Human Genome Project. With Digenome-seq it should be easier to find RNAs with fewer unintended docking sites. MORE
Image Credit: IBS and ScienceDaily.com


